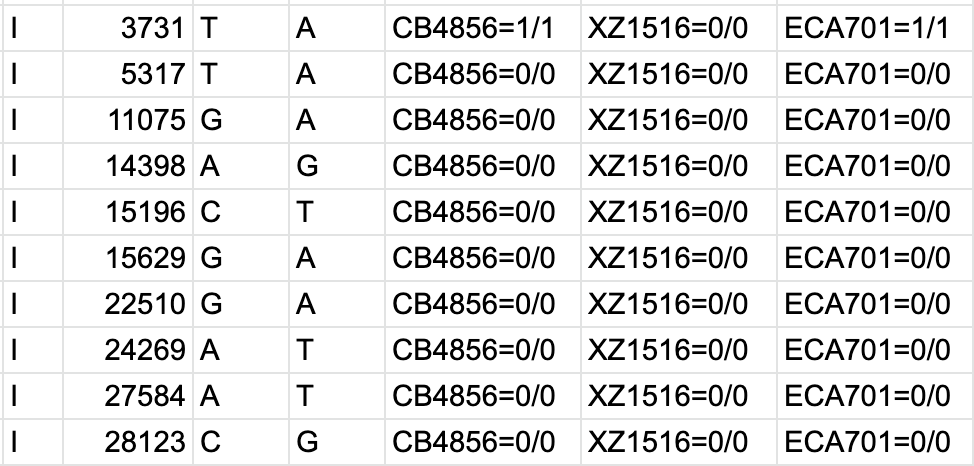
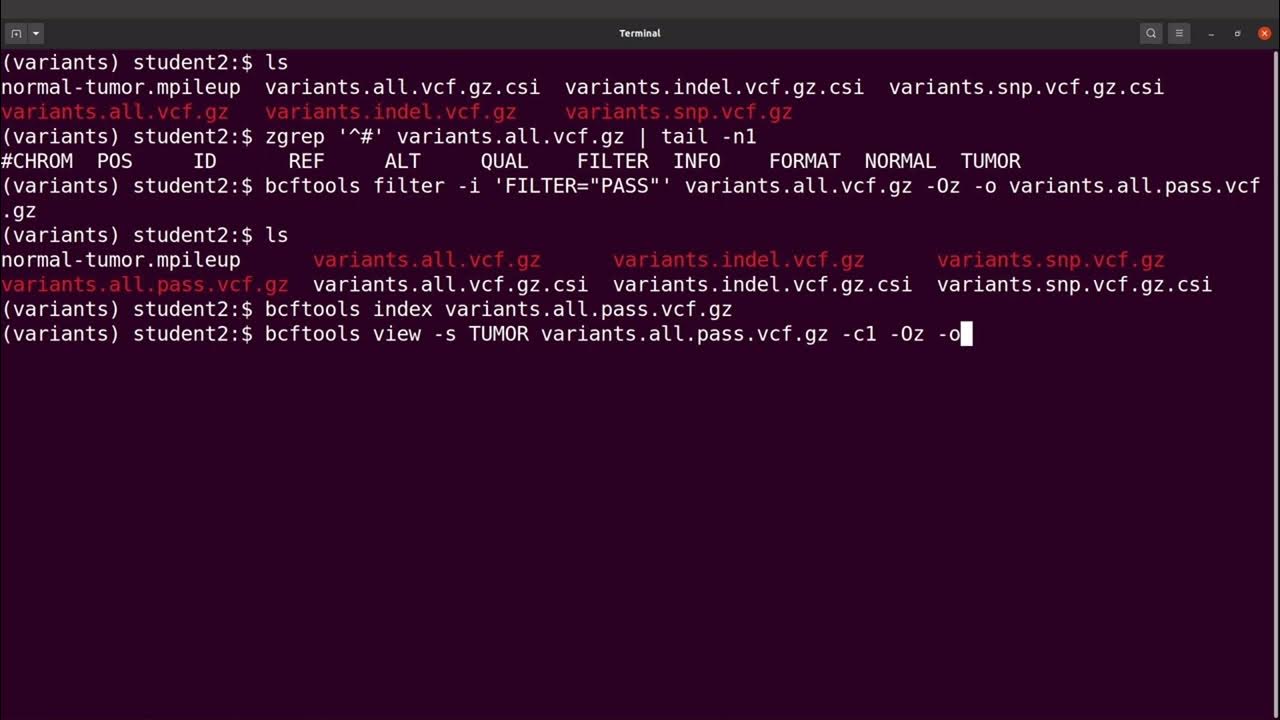
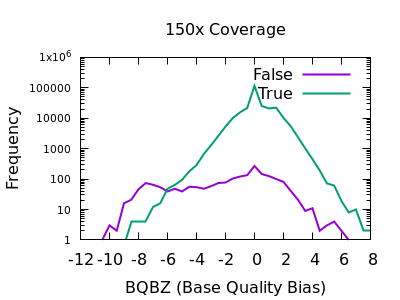
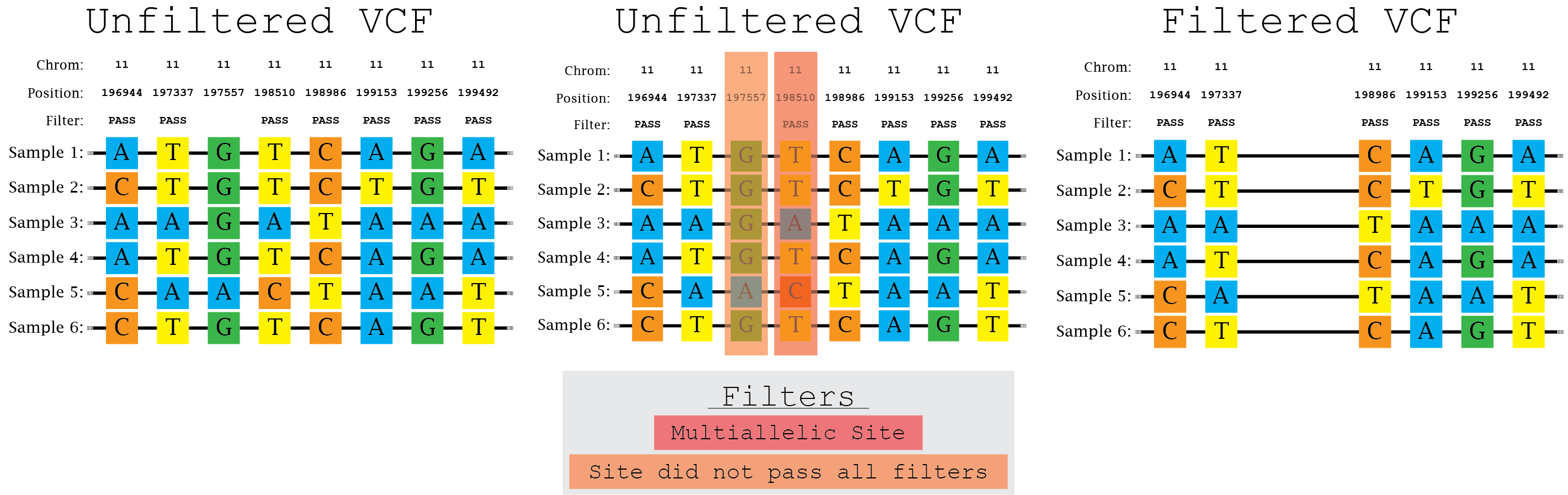
bcftools filter -e 'GT="het"' recognize GT 2/2 as heterozygous · Issue #1268 · samtools/bcftools · GitHub
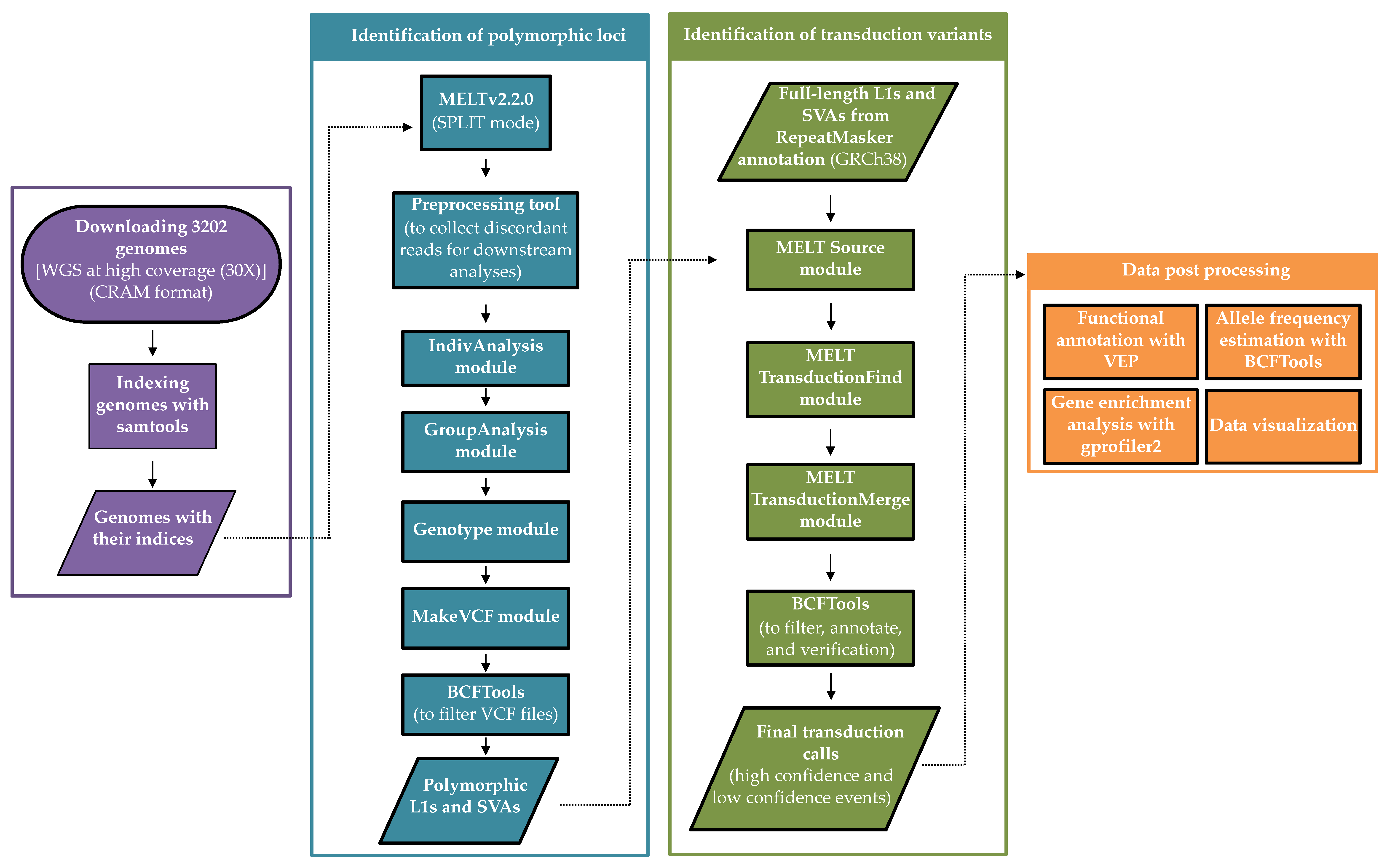
Biology | Free Full-Text | A Map of 3′ DNA Transduction Variants Mediated by Non-LTR Retroelements on 3202 Human Genomes
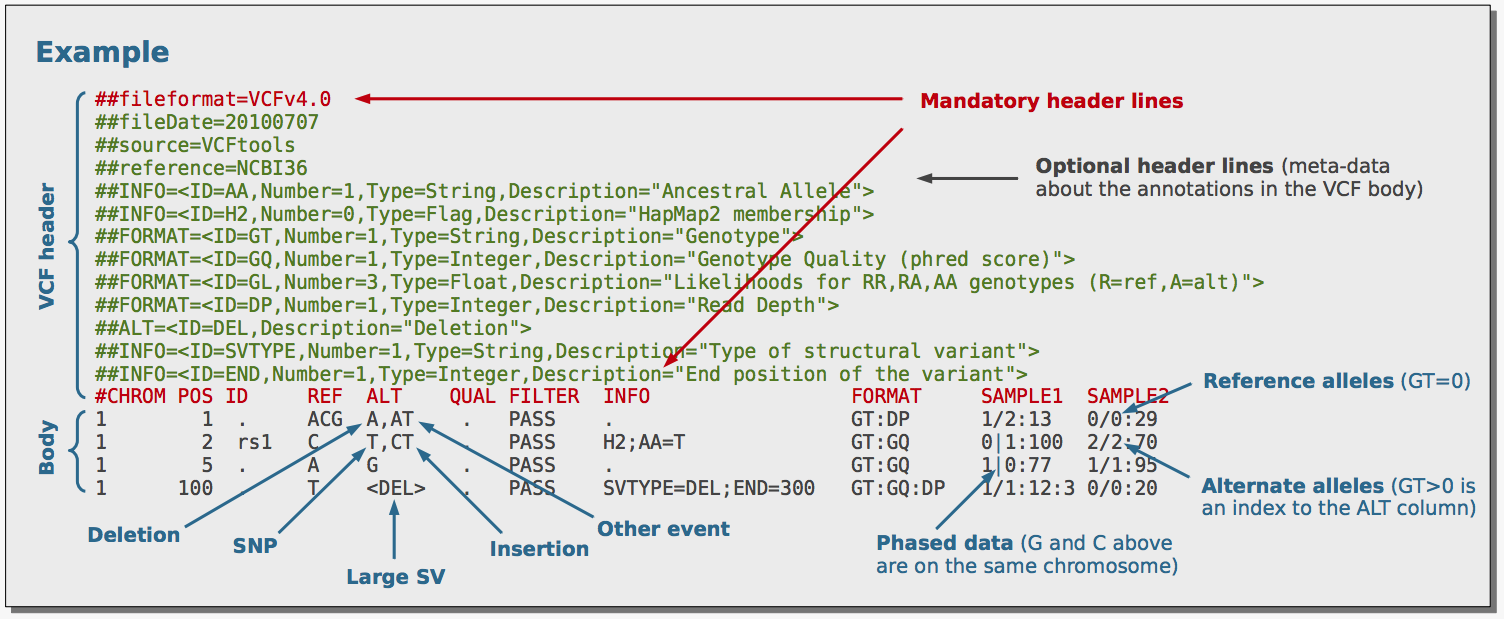
Comparing -min-DP in vcftools with filter -i 'FORMAT/DP>10' in bcftools · Issue #1384 · samtools/bcftools · GitHub
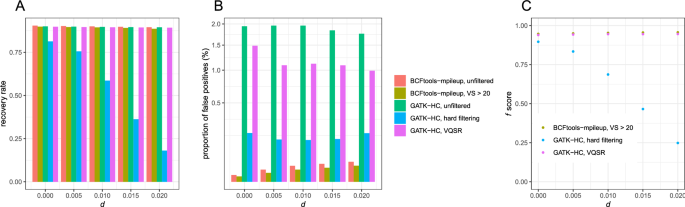
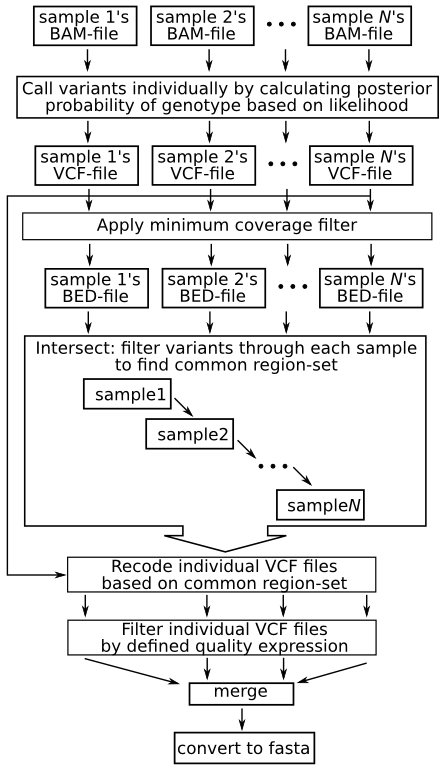
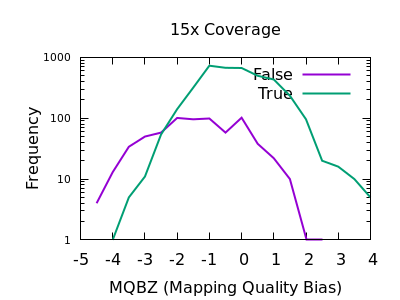
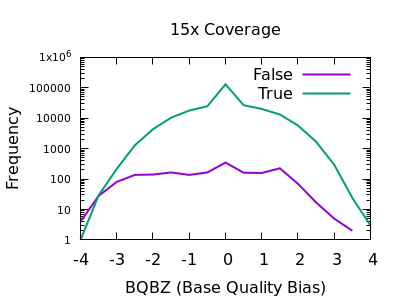
The evaluation of Bcftools mpileup and GATK HaplotypeCaller for variant calling in non-human species | Scientific Reports

The evaluation of Bcftools mpileup and GATK HaplotypeCaller for variant calling in non-human species | Scientific Reports
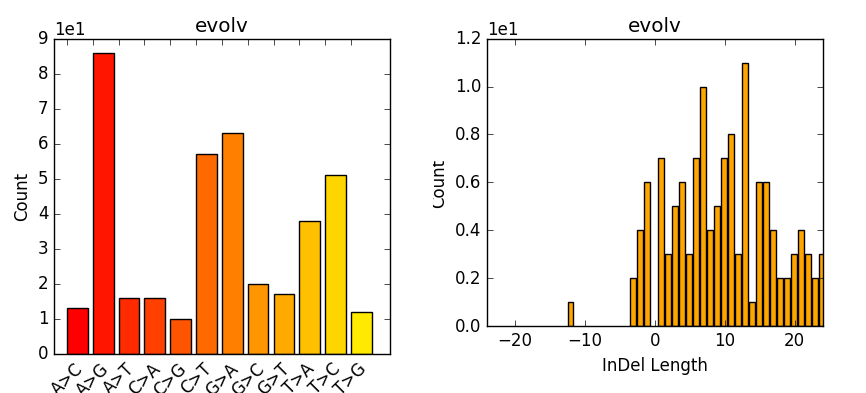
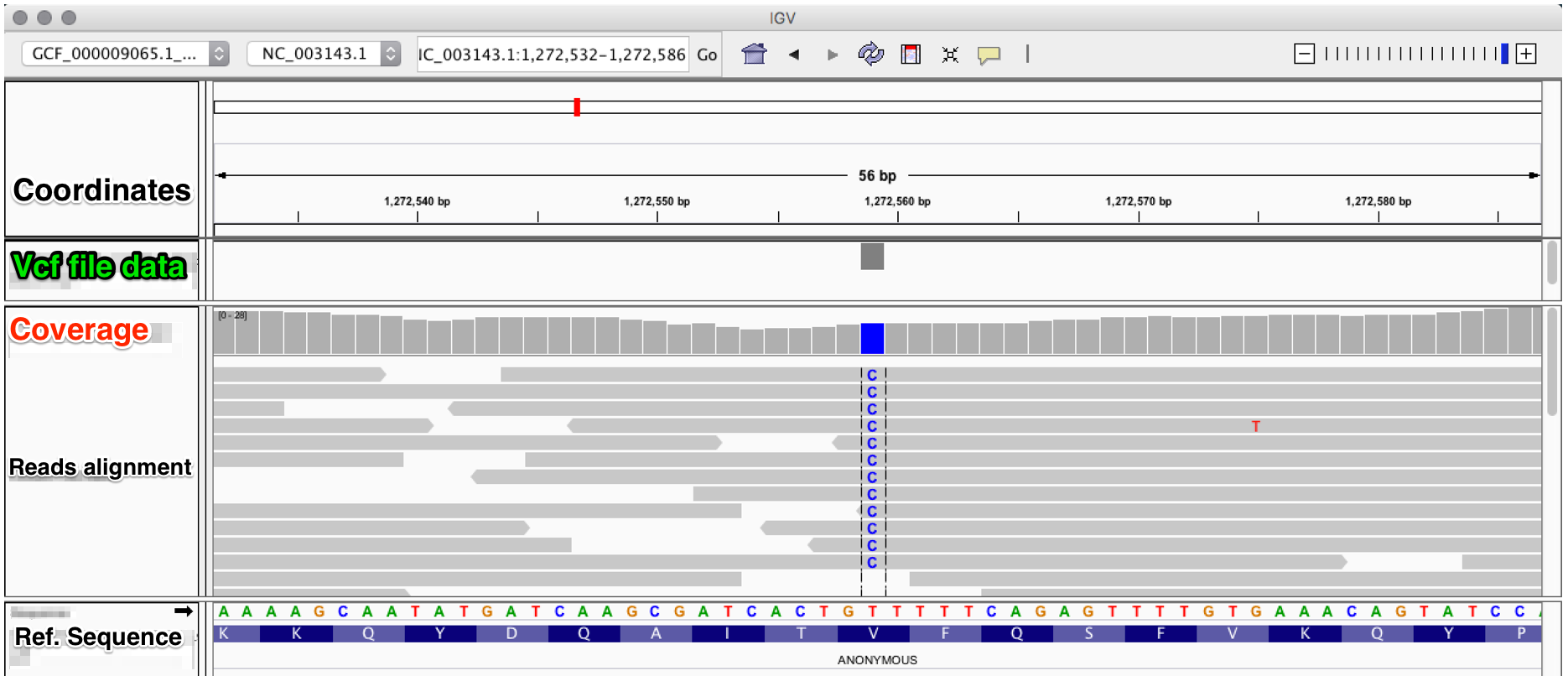
a) Filtering different variant callers VCF output for SARS-CoV-2 data.... | Download Scientific Diagram